Gram negative bacteria Type 4 Secretion System (T4SS) by Dr. Gabriel Waksman of the Institute of Structural and Molecular Biology, University College London and Birkbeck. Animations by Cellscape.
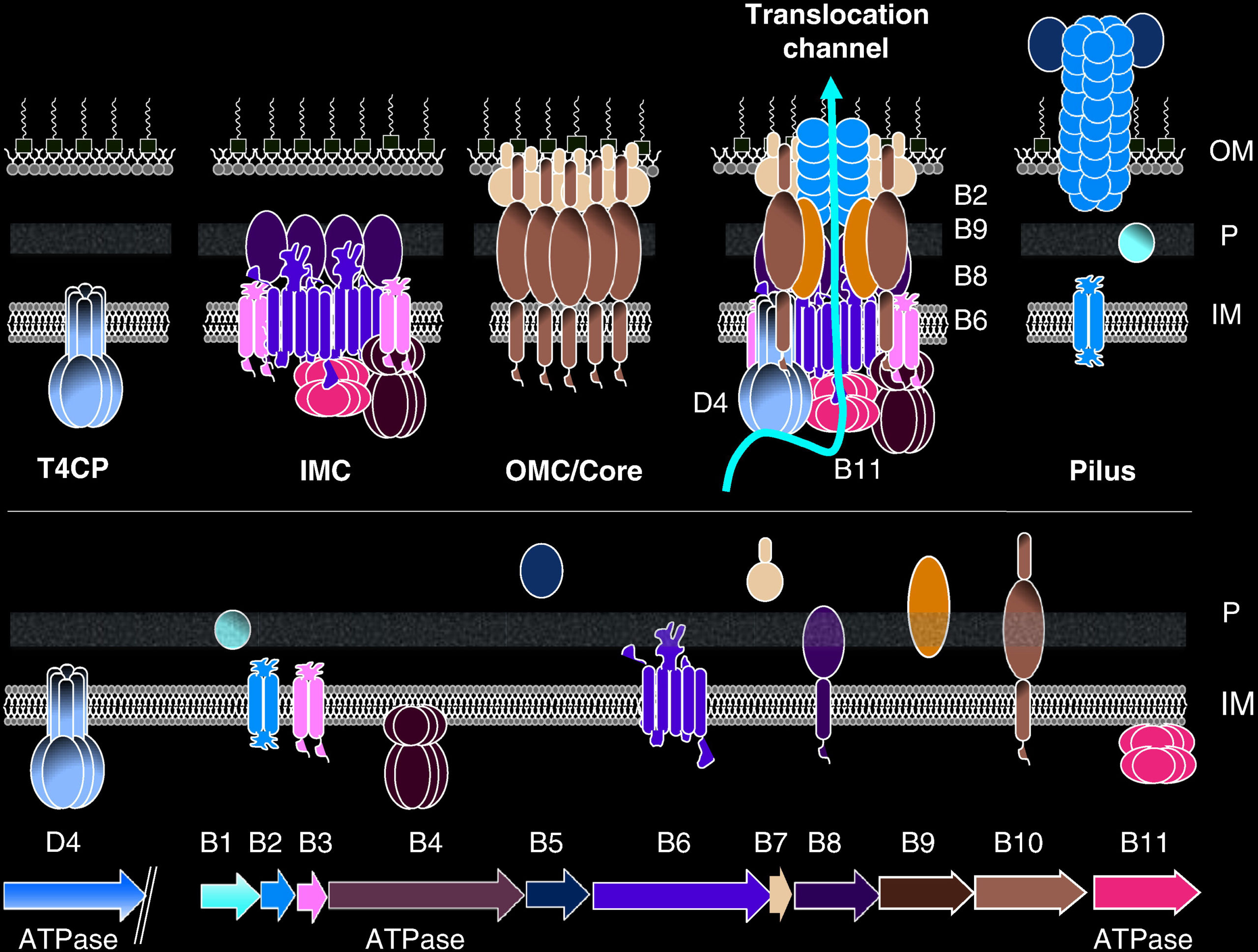
This playlist consists of three videos illustrating the type 4 secretion system (T4SS) assembly, pilus biosynthesis and transfer of DNA segment into a recipient cell. T4SS are bacterial macromolecule secreting structures. These macromolecules include proteins and DNA. Bacterial cell-to-cell exchange of genetic materials such as plasmids or mobile genetic elements is called conjugation. T4SS are composed of 12 subunits named VirB1-11 and VirD4. Three components, VirB7, VirB9, and VirB10, form the outer membrane core complex (OMCC). The OMCC connects to an inner-membrane complex (IMC) composed of VirD4, VirB4, VirB3, VirB6, VirB8 and part of VirB10. How OMCC and IMC are connected is structurally unknown. The connecting parts could be made of VirB2, VirB5, or VirB10. At least two ATPases (VirB4 and VirD4), sometimes three (VirB4, VirD4, and VirB11) power the system. In this movie, you will see the progressive assembly of the various parts of the T4SS starting with the assembly of the OMCC and then that of the IMC.
Conjugation is one of the mechanisms by which antibiotic resistance genes spread among bacterial populations. The conjugative pilus is largely made of VirB2. VirB5 also takes part in pilus assembly. Pili serves either as attachment structure to recipient cells and as a conduit for relaxase/ssDNA transport.
The following movie shows a structural mechanism for T4SS pilus biogenesis. The protein VirB2 is located in the inner membrane (IM) of the bacterium. Thousands of VirB2 units are loaded from the IM and assembled into a 5-start helical pilus array. VirB2 is not alone but is bound to a phospholipid (PL). Presumably with the aid of the ATPase VirB4, VirB2-PL complexes are extracted from the IM in large numbers to assemble the pilus. As the pilus elongates, it contacts with a recipient cell, which will trigger its retraction. Retraction of the pilus brings the the donor and the recipient cells next to each other.
During conjugation, the DNA is transferred from the donor to the recipient cell through a single-strandDNA (ssDNA) and protein conglomerate. Indeed prior to DNA transfer, a large cytosolic complex needs to be assembled around the section of the plasmid to be transferred. The region of the plasmid DNA where the complex assembles is called Origin of Transfer (OriT). The complex is called “the relaxosome” and is composed of a major protein, called “relaxase” (TraI) and some accessory proteins (TraM, TraY, and IHF). The movie below shows TraI, TraM, TraY and IHF assembling at OriT. Once the relaxosome is assembled, it will connect with the T4SS.
Upon pilus contact with a recipient cell, the relaxosome is activated. This means that the relaxase will react with the DNA in a site within OriT called “nic”. The relaxase will nick the “nic” site on the strand to be transferred. The resulting 5’phosphate from the nicking reaction will react covalently with the relaxase. The covalently interacting complex of relaxase and ssDNA that will get transported through the T4SS. Activation of the relaxosome also result in a ssDNA bubble onto which a second molecule of TraI (the relaxase) wil bind, this time through its helicase domains (in green and blue). The second TraI will be used to pump the relaxase-ssDNA through the T4SS into the recipient cell as an unfolded peptide. The peptide pumping action is presumably carried out by the VirB4 ATPase. The transport of the ssDNA, presumably driven by the VirD4 ATPase. In the movie, the switching of the relaxase-ssDNA substrate from VirB4 to VirD4 is hypothesized to be after relaxase transport is completed.


0 Comments
You can be the first one to leave a comment.