According to the Center for Disease Control (CDC) every year, 776,000 people in the United States get new herpes infections. Genital herpes infection is common in the United States. Nationwide, 15.5 % of persons aged 14 to 49 years have HSV-2 infection. The virus is also known as common cold sore. Seeing is believing. Researchers from UNC (University of North Carolina at Chapel Hill) have visualized visualized the structure and action of a key protein in this sexually transmitted virus: the Infected Cell Protein 8 (ICP8).
The animation was prepared by Manuel Esguerra, Ph.D. and Gökhan Tolun, Ph.D. who is the first author of a manuscript published in Nucleic Acids Research. Tolun was able to provide a new view of how such large protein machines can facilitate DNA annealing and recombination. The image Tolun created is a blend of his skill in computer image reconstructions and his hobby of photography. He is one of the few scientists who can communicate science through art. The visualization is among the winners of the 2014 BioArt Scientific Visualization Contest.
Tolun found electron microscopy images prepared earlier by Alexander Makhov, Ph.D., of the University of Pittsburgh School of Medicine and wanted to reconstruct these images in 3-D. Tolun had to master specialized scientific software packages used to determine the 3-D structures of the biological complexes. It also got featured in the blog of the NIH Director Francis Collins.
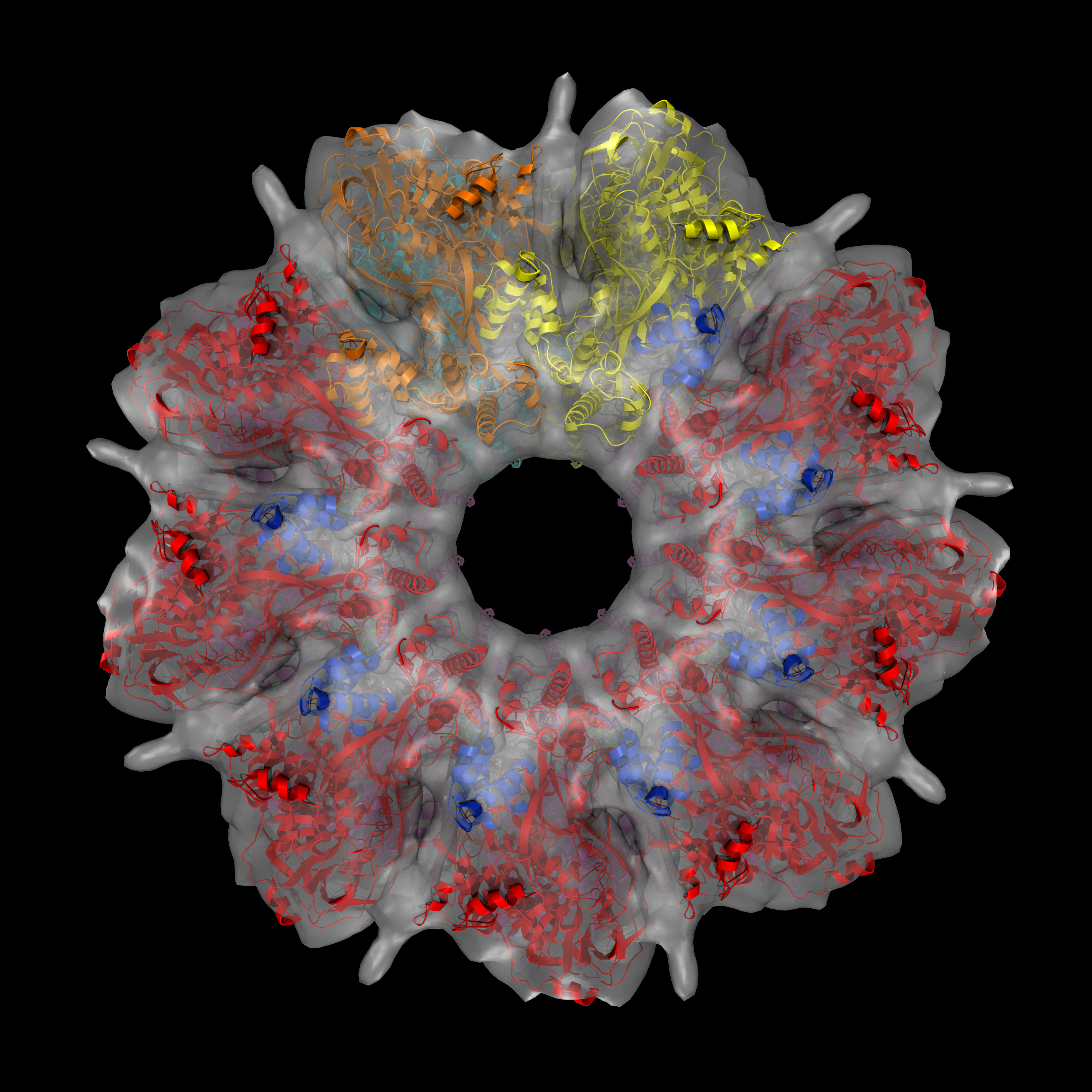
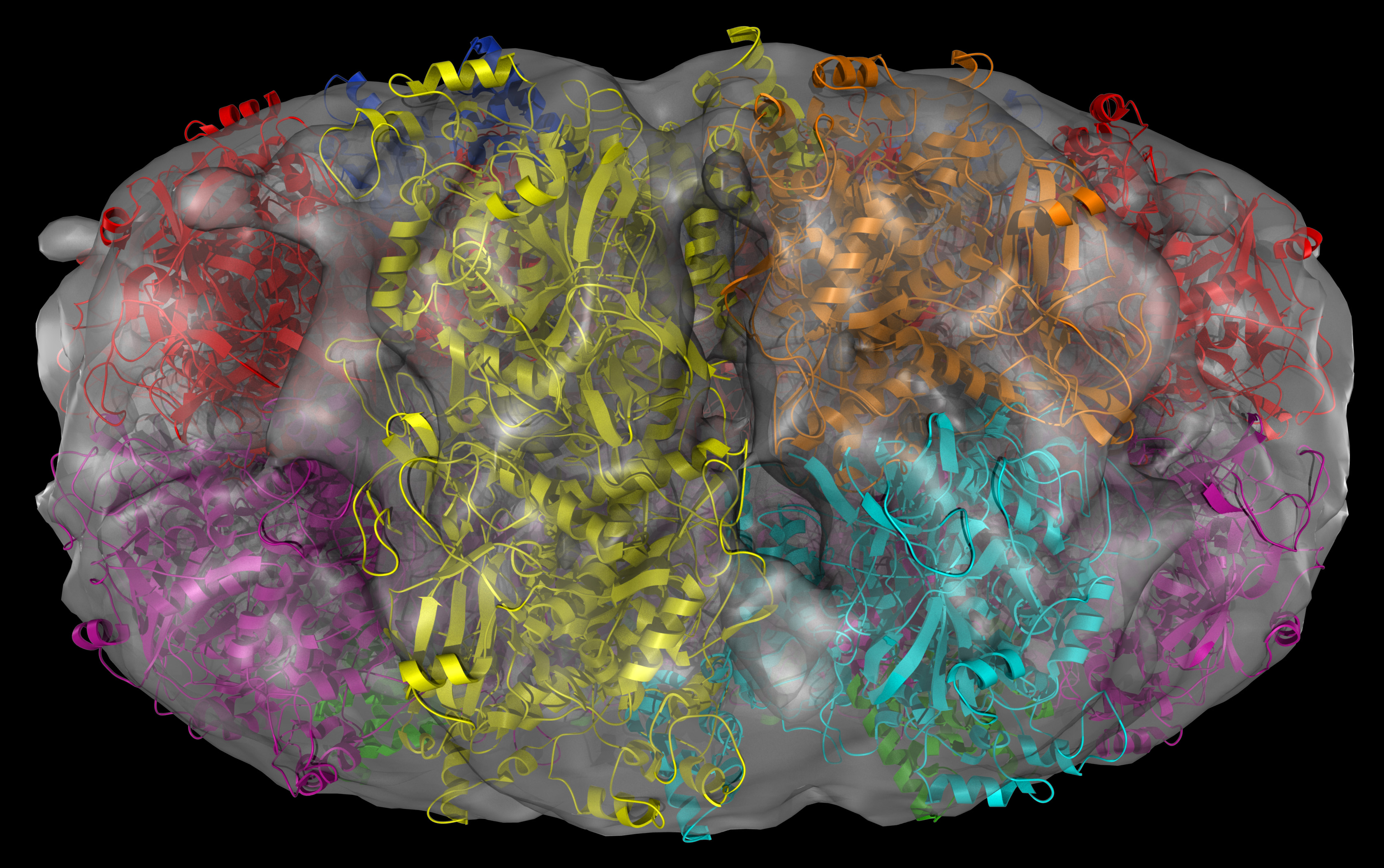
Indeed, this is a “protein machine” made of multiple parts forming rings. There is so much happening in the 18 seconds long animation. Briefly, the animated model shows single strands of DNA (red), bound circularly to each half of the ICP8 protein (bluish purple). The two halves come together to assemble a protein scaffold complex. This complex helps the single stranded DNA strands to align with each other and form the classic double stranded DNA that everyone is familiar with. You can check out the details of the protein structure below from multiple angles. Note that colors are different from that of the animation:
In this protein structure eighteen copies of ICP8 monomers (nine on each ring) come together to form two rings. Red and magenta colors are the N-terminal domains while blue and green are the C-terminal domains for each ICP8 monomers. Cyan and orange monomers were colored such that to show individual monomers with their N- and C-termini.
Science can be beautiful and art can be informative. If you further want to satisfy the bio-geek within you here are the technical details of this impressive protein machine outlined in the abstract of the study:
The Infected Cell Protein 8 (ICP8) from herpes simplex virus 1 was first identified as a single-strand (ss) DNA-binding protein. It is essential for, and abundant during, viral replication. Studies in vitro have shown that ICP8 stimulates model replication reactions, catalyzes annealing of complementary ssDNAs and, in combination with UL12 exonuclease, will catalyze ssDNA annealing homologous recombination. DNA annealing and strand transfer occurs within large oligomeric filaments of ssDNA-bound ICP8. We present the first 3D reconstruction of a novel ICP8-ssDNA complex, which seems to be the basic unit of the DNA annealing machine. The reconstructed volume consists of two nonameric rings containing ssDNA stacked on top of each other, corresponding to a molecular weight of 2.3 MDa. Fitting of the ICP8 crystal structure suggests a mechanism for the annealing reaction catalyzed by ICP8, which is most likely a general mechanism for protein-driven DNA annealing.



0 Comments
You can be the first one to leave a comment.