This unnarrated molecular animation by Alan Cheung and Patrick Cramer details the first step of the Central Dogma of biology where the messenger RNA becomes synthesized from it’s DNA template. Enzymes that read information on DNA and produce the RNA counterparts are known as RNA polymerases. There are three types of RNA polymerase enzymes (RNA polymerase I, II and III). Here the working mechanism of type II that reads protein coding genes is shown based on multiple scientific studies spanning decades of intense experimental effort.
Transcription of a gene into a messenger RNA requires transcription factors. DNA binding transcription factor proteins are the key initiators of this process: they mark the beginning of the gene for RNA Polymerase II just like the approach lights at an airport runway.
RNA Polymerase II is a large enzyme that has 10 “core” subunits. On top of the 10 subunit Polymerase II core two more subunits called Rpb4/7 subcomplex join and altogether form the complete, 12 subunit enzyme. All structures shown are modeled around the Yeast 12 subunit RNA Polymerase II (1WCM).
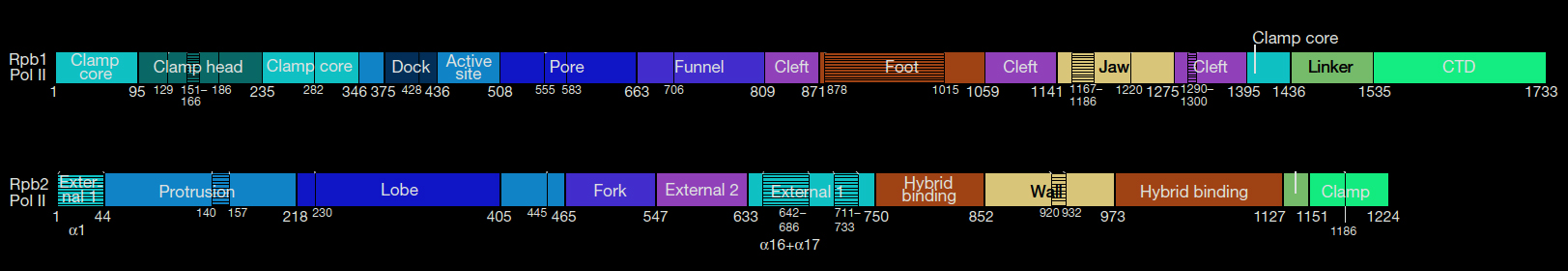
The two largest subunits (RPB1 and RPB2) of the RNA Polymerase II have domains that enable production of RNA from a DNA template. These sections or domains are named as Clamp, Dock, Active site, Pore, Funnel, Cleft, Foot, Jaw, Expander, Hybrid binding, Wall, Protrusion. The largest subunit RPB1 also has an additional C-terminal Domain (CTD) which is not shown on the 3-D molecular model. CTD is an exceptionally floppy whip-like structure hard to show in a static conformation. Yeast and Human RNA polymerase II have been well studied.
The movie starts with the formation of RNA Polymerase II-TFIIF (Transcription Factor F) complex. Transcription initiation begins with the formation of a closed promoter complex, which contains the 10 subunit Pol II core, the Pol II subcomplex Rpb4/7, and the transcription factors (TF) IID (which includes the TATA-box binding protein TBP and TBP-associated factors), TFIIB, TFIIE, TFIIF, and TFIIH. Rpb4/7 binding stabilizes a closed conformation of the Pol II clamp domain, which only permits passage of single-stranded DNA to the active site. Subsequent binding of TFIIF to Pol II generates the complete Pol II-TFIIF complex. The dimerization domain of TFIIF is positioned on the lobe domain of Pol II.
In the next stage of the movie, the Pol II-TFIIF complex binds onto the TBP-TFIIB-DNA complex, to form a closed promoter complex. The closed complex model was derived by combining crystal structures of the Pol II-TFIIB complex and the TBP-TFIIB-DNA complex. This model reveals the central role of TFIIB as a bridge between the promoter and the polymerase. Docking of the TBP-TFIIB-DNA complex onto the Pol II-TFIIF complex involves the binding of the TFIIB N-terminal ribbon domain to the Pol II dock domain and the binding of the C-terminal TFIIB core domain to the polymerase wall. The TFIIB reader and linker regions connect the N- and C-terminal domains of TFIIB and extend through the Pol II cleft. The entire initiation complex looks like the following animation:
Transition from the closed to the open promoter complex happens by separation of the DNA strands to form an unwound DNA region. Once the transcription ‘‘bubble’’ forms the template single strand DNA gets positioned in the active center of Pol II. RNA synthesis then can initiate from the transcription start site. The initially transcribing complex (ITC) is unstable and releases short RNAs during abortive initiation (not shown in the movie). When the RNA reaches a certain length, initiation factors are released, and a stable elongation complex (EC) is formed. Elongation complex contains a DNA-RNA hybrid of eight to nine base pairs. During transcription elongation, the EC repeatedly performs the nucleotide addition cycle (NAC) to attach a nucleotide to the growing messenger RNA (mRNA) chain by catalyzing DNA template-directed formation of an RNA phosphodiester bond.
Errors do occur during RNA transcription and must be corrected to prevent synthesis of mutated, nonfunctional proteins that possibly may lead to cell death. When an incorrect base is incorporated to a growing RNA chain, the RNA–DNA helix distorts. The distortion of the helix stalls the movement of RNA Polymerase II and the enzyme pulls over into a “backtracked” state waiting for the wrong nucleotide to be removed from the strand. Molecular proofreading ability of RNA polymerases minimizes transcription errors ensuring accuracy in the transmission of the genetic information from DNA to the proteins that allow our bodies to function. Elongation factors, such as TFIIS and Spt4/5, are required to deal with these obstacles. TFIIS can reactivate an arrested EC by stimulating RNA cleavage, and Spt4/5 can keep polymerase to remain bound to the DNA.


1 Comment