The Inner Life of the Cell is a 3D computer graphics animation by David Bolinsky, former lead medical illustrator at Yale. He collaborated with lead animator John Liebler, and Mike Astrachan at XVIVO for this production. It illustrates the molecular interactions that occur when a white blood cell (Leukocyte) in the blood vessels of the human body is triggered by inflammation. This is also called leukocyte extravasation. It begins with a white blood cell rolling along the inner surface of the capillary veins. Then we get into details of molecular structures both outside and inside the leukocyte. In the end, the leukocyte changes its overall shape to pass through the cells of the capillary vein wall in order to reach the inflammation to initiate the immune reaction.
The Inner Life of Cell is very dense in terms of information content. At the same time, it is a companion resource for much later productions such as BBC’s “The Hidden Life of Cell” made in 2012. So here’s a summary trying to keep jargon at minimum:
Cell adhesion proteins (P-selectins) on surfaces of the cells lining the capillary veins interact with other proteins called PSGL1 found on white blood cell surface. These two proteins cover the surfaces of both cell types. Leukocytes can roll on surfaces of the capillary veins like balls covered with velcro. Existing interactions among P-selectins and PSGL1 are broken while new ones are formed leading to locomotion like the belt of a crawler tractor. Understanding the environment outside of animal cells gives us an idea about how cells interact among themselves.
Cells are bounded by membranes made of lipid bilayer. Layers are called leaflets. The outer leaflet of the lipid bilayer is enriched in sphingolipids, and phosphatidyl choline. Sphingolipid rich sections stand out above the rest of the leaflet. These raised platforms that look like rafts attract certain types of membrane proteins. Rigidity of these lipid rafts depends on cholesterol molecules. Outside the rafts, bends in unsaturated hydrocarbon chains and lower cholesterol concentration leads to increased fluidity of the membrane.
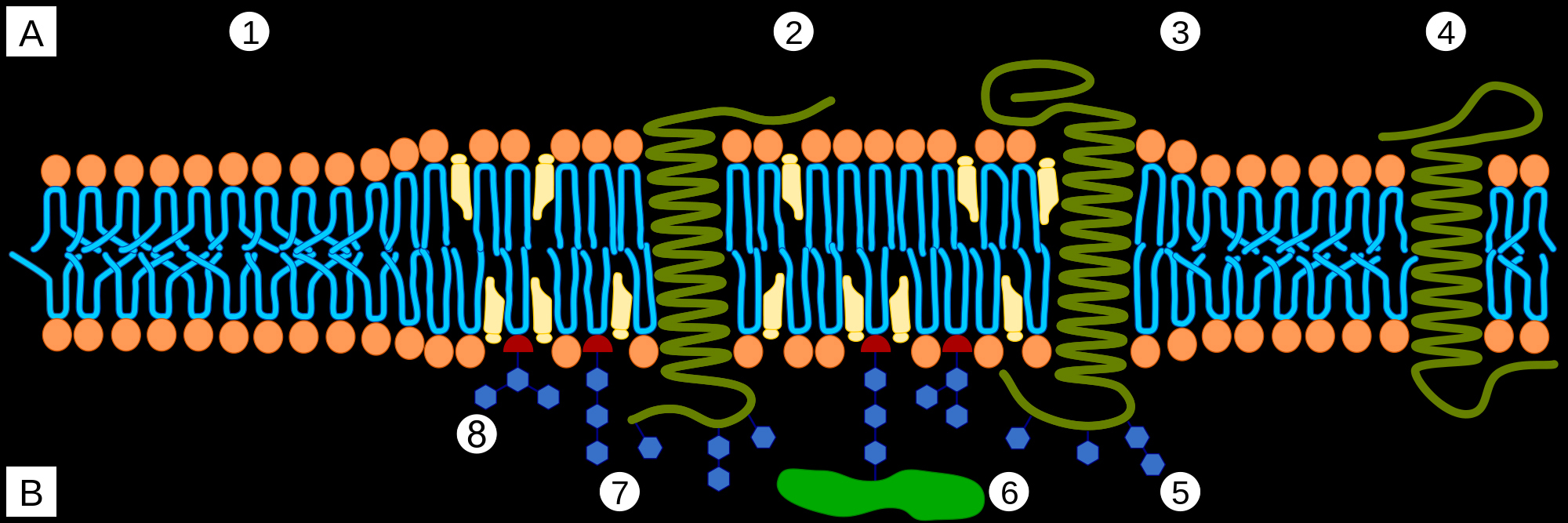
A Intracellular space or cytosol
B Extracellular space or vesicle/Golgi apparatus lumen
1. Non-raft membrane
2. Lipid raft
3. Lipid raft associated transmembrane protein
4. Non-raft membrane protein
5. Glycosylation modifications (on glycoproteins and glycolipids)
6. GPI-anchored protein
7. Cholesterol
8. Glycolipid
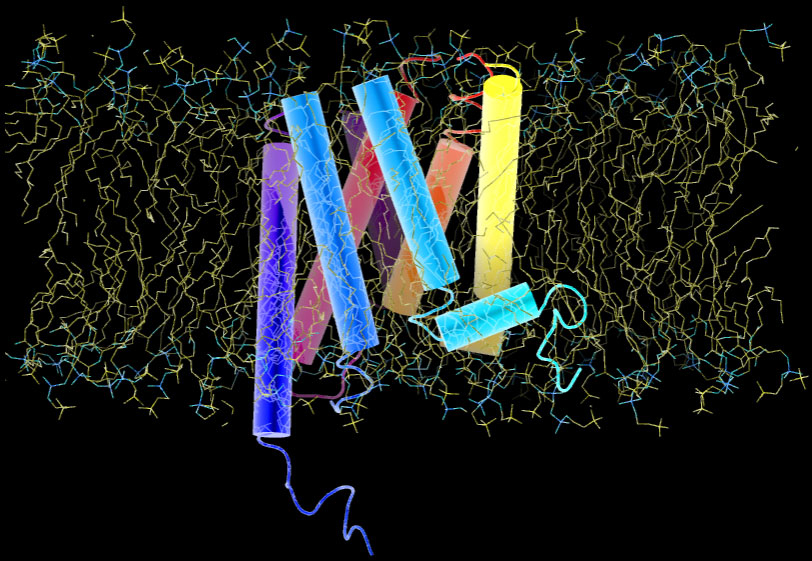
At sites of inflammation, a group of signalling proteins called chemokines are secreted. Chemokines bound to heparan sulfate proteoglycan on capillary vein cell surfaces, interact with leukocytes seven transmembrane receptors. The binding stimulates leukocytes and triggers a series of signaling reactions inside the cell. Below is a representation of a G-protein receptor with 7 transmembrane domains that look like dynamite sticks. The entire receptor is embedded in lipid bilayer:
The chemical composition of inner and outer leaflets of the lipid bilayer differs. As we see in the 7TM protein above, some proteins traverse the membrane, others are partially embedded or rooted into the inner leaflet or are held by other membrane proteins. Transmission of signals across the cell membrane relies on these proteins.
Beneath the lipid bilayer, Spectrin tetramers, arranged into a hexagonal network, are anchored by membrane proteins. This network forms the membrane skeleton that contributes to membrane stability, and membrane protein distribution.
The cytoskeleton is a network of filamentous proteins that are responsible for organization of cytosolic components in space. Leukocytes attack foreing bodies using projections called microvilli. Inside microvilli, actin filaments form tight bundles which are held by cross-linking proteins.
Filaments, capped at their minus ends by a protein complex, grow away from the plasma membrane by the addition of actin monomers to their plus end. The actin network is a very dynamic structure, with continuous directional elongation by polymerization and shortening by disassembly. Severing proteins induce bends in the filament and lead to the formation of short fragments that rapidly disintegrate, or give rise to new filaments. The cytoskeleton includes a network of microtubules created by the lateral association of proto-filaments formed by the polymerization of tubulin dimers.
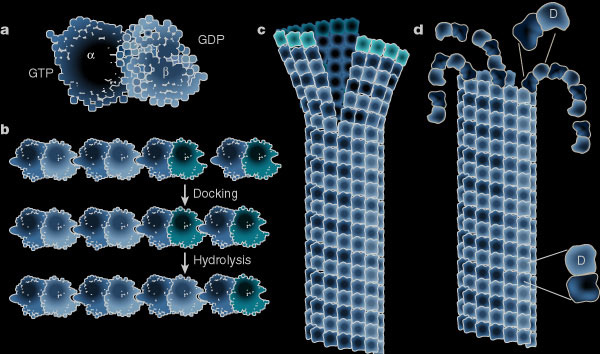
While the plus ends of some microtubules extend towards the plasma membrane, proteins stabilize the curved confirmation of proto-filaments from other microtubules causing their rapid plus end depolymerisation. Microtubules provide tracks along which membrane-bound vesicles travel to and from the plasma membrane. The following illustration shows structural changes in the microtubule as it grows and shrinks like miltiple zippers operating at the same time. (a) Molecular structure of the tubulin dimer forming the wall of the protofilament. (b) Docking of the a-b subunit to the microtubule end. Residues from the incoming a-subunit trigger hydrolysis of the GTP bound to the lattice attached to b-subunit. (c) – (d) Microtubules at growing ends contain sheets of protofilaments while microtubules at shrinking ends curl. The straight–bent transition is also shown in (d). The GTP dimer has a straight conformation that fits nicely into the straight wall of the microtubule. Hydrolysis of GTP induces a bend in the subunit which places stress on the lattice, allowing the protofilament to curl outwards.
The directional movement of membrane-packaged cargo vesicles is fascilitated by a family of motor protein linking vesicles and microtubules. Membrane-bound organelles like mitochondria swims through the cytoskeleton like a bacteria. While doing so, mitochondria change their shapes continuously and interact with microtubules.
All the microtubules originate from the centrosome, a discreet fibrous structure containing two centrioles oriented perpendicular to each other. Centrioles and located near the cell nucleus.
Nuclear pore complexes which serve as selective gates between the nucleus and the cytoplasm recognize nuclear export signal carrying proteins and let messenger RNA exit into the cytoplasm. The following video provides a structural rendering of the Nuclear Pore Complex produced by Samir S. Patel of the Rexach Lab from University of California Santa Cruz:
Into the Nucleus from Nature Documentaries on Vimeo.
Nucleus is where the Central Dogma of biology starts. Information stored in DNA is copied into messenger RNA (mRNA) molecules. These mRNAs exit the nucleus through nuclear pores into the cytoplasm. In cytoplasm mRNA molecules form a translation ring. Floating ribosomes binds onto the rings and translate the circularized mRNA molecules into proteins. Some of these proteins reside in the cytosol while others get imported into mitochondria or other organelles. If the synthesised proteins will be secreted outside the cell the free floating ribosomes dock on to protein translocators at the surface of the endoplasmic reticulum (ER). Newly synthesized proteins pass through a pore in the translocator. Cell secreted proteins accumulate inside the endoplasmic reticulum. Similarly, integral membrane proteins get embedded in the endoplasmic reticulum membrane.
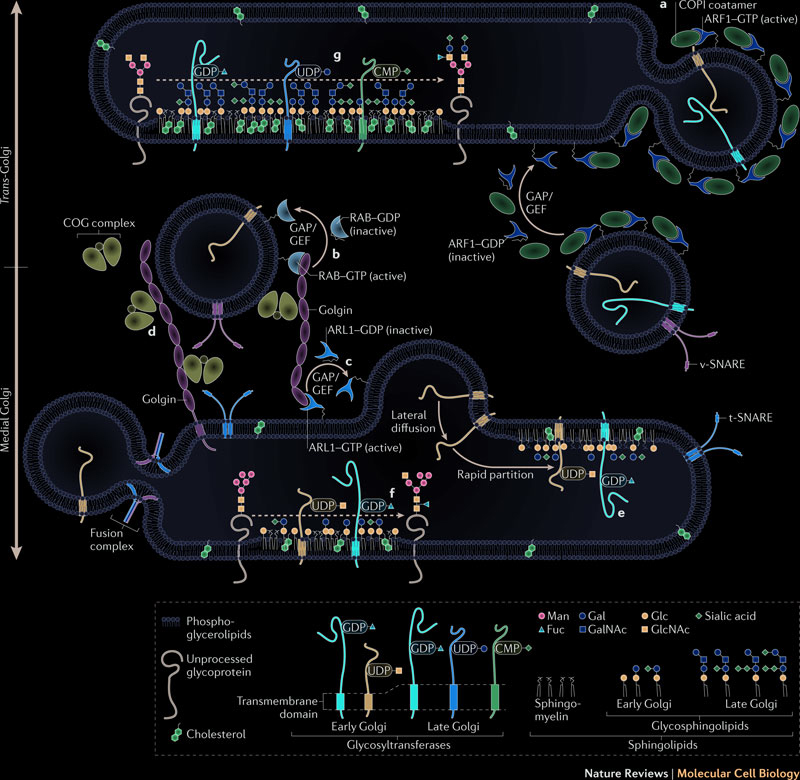
Proteins are transported from the endoplasmic reticulum to the Golgi apparatus by vesicles traveling along the microtubules. Protein glycosylation, initiated in the endoplasmic reticulum, is completed inside the Golgi apparatus. Fully glycosylated proteins are transported from the Golgi apparatus to the plasma membrane. When a vesicle fuses with the plasma membrane, proteins inside the vesicles are secreted, and proteins embedded in the vesicles membrane diffuse in the cell membrane. Protein glycosylation is a very dynamic and complex process which modifies and makes proteins ready for diverse functions. The following illustration summarizes the traffic between endoplasmic reticulum and golgi:
At sites of inflammation, chemokines secreted by cells lining the capillary veins, bind to the sections of G-protein coupled membrane receptors sticking out of the cell membranes. This binding causes a structural change in the portion of the receptor extending towards the inside of the cell, and the the G-protein coupled receptor gets activated.
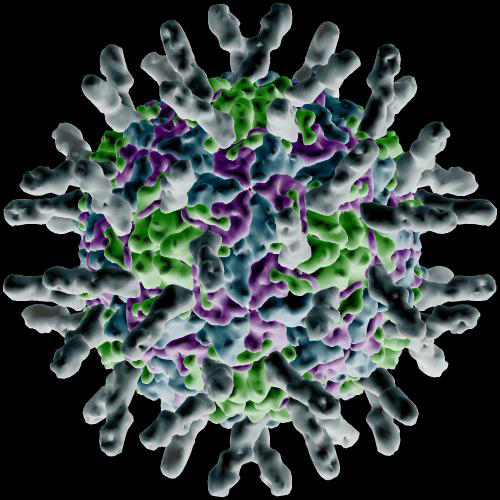
The activation of the G-proteins triggers protein activation in rapid succession, which leads integrins to gather and form lipid rafts. As the integrins come together a large structural change occurs in them allowing interaction with I-Cam proteins (Intercellular Adhesion Molecule) at the surface of the cells forming the walls of capillaries. The image below is a rendering of how I-Cam molecules (gray colored structures sticking out) surround a human Rhinovirus.
These molecular interactions hold the rolling leukocyte at the site of inflammation. Cytoskeletal reorganization results in flattening of the leukocyte. The white blood cell then penetrates between cells forming the walls of the capillary and reaches into the inflamed tissue.
Rolling, activation, adhesion, and trans-endothelial penetration are the four steps of the process called leukocyte extravasation.
Please keep in mind that all these animations are broad simplifications of actual cellular events. In reality cells are extremely crowded with molecular structures. The following video attempts to show what inner workings of a cell may look like. Structures are slowed down so that we can see their extremely fast movements. If you are interested in learning more about molecular structures in living bodies you can find more under molecular nature category.
Protein packing:


2 Comments
Dear Sir/ Madam
While wishing your success, I would like to have the full written text of The Inner Life of the Cell which is a 3D computer graphics animation by David Bolinsky, as I can not understand the speech of the narrator due to his accent .
My Eamail is : [email protected]
Much appreciation in advance.
Yours
Farghadan